We can calculate 3-dimensional surface area in ArcMap 10.x (ArcMap; Environmental Systems Research Institute, Redlands, California) using standard 30-m United States Geological Survey DEMs and the DEM Surface Tools for ArcMap extension (Jenness 2004). We acquired all DEM data from United States Department of Agriculture, Natural Resource Conservation Service (http://datagateway.nrcs.usda.gov/) at the Geospatial Data Gateway. We can then use the surface area tool to calculate true surface area of the landscape for each grid cell using the DEM elevation from the surrounding 8 cells. The new grid cell values represented the 3-dimensional surface area for the land area contained within that cell's boundaries. We then summed all grid cell values within the animal's home-range polygon to derive a topographic home range for each individual.
Alternatively, we can create home ranges in R and look at them in 3D using the package rasterVis that now includes the rgl package. We can also view DEMs in R and create slope, aspect, or hillshade using the rasterVis package.
- Exercise 6.1 - Download and extract zip folder into your preferred location
- Set working directory to the extracted folder in R under File - Change dir...
- First we need to load the packages needed for the exercise
library(rasterVis)
library(raster)
library(sp)
library(rgdal)
library(maptools)#writeAsciiGrid
library(ks)#hpikde.ud
library(adehabitatHR)
library(adehabitatMA) - Now open the script "Raster3dScript.R" and run code directly from the script
muleys<-read.csv("DCmuleysedited.csv")
str(muleys)
#Remove outlier locations
newmuleys <-subset(muleys, muleys$Long > -110.90 & muleys$Lat > 37.80)
muleys <- newmuleys
newmuleys <-subset(muleys, muleys$Long < -107)
muleys <- newmuleys
muleys$NewDate<-as.POSIXct(muleys$GPSFixTime, format="%Y.%m.%d %H:%M:%S")
#TIME DIFF NECESSARY IN BBMM CODE
timediff <- diff(muleys$NewDate)*60
# remove first entry without any difference
muleys <- muleys[-1,]
muleys$timelag <-as.numeric(abs(timediff))
#Remove locations greater than 24 hours apart in time
muleys <- subset(muleys, muleys$timelag < 18000)
#Code separate each animal into a shapefile or text file
# get input file
indata <- muleys
innames <- unique(muleys$id)
innames <- innames[1:2]
outnames <- innames
# begin loop to calculate home ranges
for (i in 1:length(innames)){
data <- indata[which(indata$id==innames[i]),]
if(dim(data)[1] != 0){
# export the point data into a shp file
data.xy = data[c("X", "Y")]
coordinates(data.xy) <- ~X+Y
sppt <- SpatialPointsDataFrame(coordinates(data.xy),data)
proj4string(sppt) <- CRS("+proj=utm +zone=12 +datum=WGS84")
#writePointsShape(sppt,fn=paste(outnames[i],sep="/"),factor2char=TRUE)
sppt <-data[c(-8,-9)]
write.table(sppt, paste(outnames[i],"txt",sep="."), sep="\t", quote=FALSE,
row.names=FALSE)
write.table(paste(outnames[i],"txt",sep="."), "In_list.txt",sep="\t",
quote=FALSE, row.names=FALSE, col.names=FALSE, append=TRUE)
}} - Reads in each text file for each animal using the In_list.txt file
List<-read.table("In_list.txt",sep="\t",header=F)
head(List) #"List" contains the filenames (e.g. "D4.txt") of the deer data
# sets exported from code above - Begin loop to create home ranges for each deer using hplug-in
for(i in 1:nrow(List)) {
coords<-read.table(as.character(List[i,]),sep="\t",header=T)
loc<-coords[,c("X", "Y")]
#Reference grid : input parameters
RESO <- 100 # grid resolution (m)
BUFF <- 5000 # grid extent (m) (buffer around location extremes)
XMIN <- RESO*(round(((min(coords$X)-BUFF)/RESO),0))
YMIN <- RESO*(round(((min(coords$Y)-BUFF)/RESO),0))
XMAX <- XMIN+RESO*(round(((max(coords$X)+BUFF-XMIN)/RESO),0))
YMAX <- YMIN+RESO*(round(((max(coords$Y)+BUFF-YMIN)/RESO),0))
NRW <- ((YMAX-YMIN)/RESO)
NCL <- ((XMAX-XMIN)/RESO)
#Generation of refgrid
refgrid<-raster(nrows=NRW, ncols=NCL, xmn=XMIN, xmx=XMAX, ymn=YMIN, ymx=YMAX)
refgrid<-as(refgrid,"SpatialPixels")
#str(refgrid)
#PKDE computation
#Convert the SpatialGrid class to a raster
sampRaster <- raster(refgrid)
##set all the raster values to 1 such as to make a data mask
sampRaster[] <- 1
##Get the center points of the mask raster with values set to 1
evalPoints <- xyFromCell(sampRaster, 1:ncell(sampRaster))
##Here we can see how grid has a buffer around the locations and ##trajectory.
##This will ensure that we #project our home range estimates into a ##slightly larger extent than the original points extent
(bbox) alone.
#plot(sampRaster)
#lines(loc, cex=0.5, lwd=0.1, col="grey")
#points(loc, pch=1, cex=0.5)
##Calculate Hpi from the xy coordinates we used above, Hpi performs bivariate
## smoothing whereas hpi
#performs univariate. Bivariate is preferred.
Hpi1 <- Hpi(x = loc)
##write out the bandwidth matrix to a file as you might want to refer to it ##later
#write.table(Hpi1, paste("hpivalue_", range, ".txt", sep=""), row.names=FALSE, sep="\t")
##Create the KDE using the evaluation points
hpikde <- kde(x=loc, H=Hpi1, eval.points=evalPoints)
##Create a template raster based upon the mask and then assign the ##values from the kde to the template
hpikde.raster <- raster(refgrid)
hpikde.raster <- setValues(hpikde.raster,hpikde$estimate)
##We can take this raster and put it back into an adehabitatHR object
##Cast over to SPxDF
hpikde.px <- as(hpikde.raster,"SpatialPixelsDataFrame")
##create new estUD using the SPxDF
hpikde.ud <- new("estUD", hpikde.px)
##Assign values to a couple slots of the estUD
hpikde.ud@vol = FALSE
hpikde.ud@h$meth = "Plug-in Bandwidth"
##Convert the UD values to volume using getvolumeUD from ##adehabitatHR and cast over to a raster
udvol <- getvolumeUD(hpikde.ud, standardize=TRUE)
#Write each home range to an ascii file if needed.
#writeAsciiGrid(udvol, paste(substr(List[i,],1,7),"PKDE","asc", sep=".")) - Generate 3D of each home range to plot in separate windows all within the home range loop.
if (require(rgl)) {
#data(loc)
r <- raster(udvol)
extent(r) <- c(0, 610, 0, 870)
drape <- cut(r, 5)
plot3D(r, drape=drape, zfac=2)
}} - We can also run code for creating KDE using href smoothing for comparison
#Begin loop to generate home ranges
for(i in 1:nrow(List)) {
coords<-read.table(as.character(List[i,]),sep="\t",header=T)
head(coords)
loc<-coords[,c("X", "Y")]
coordinates(loc) = c("X", "Y")
#Coordinate system info may not be needed
proj4string(loc) = CRS("+proj=utm +zone=12 +datum=WGS84")
#Generation of a reference grid around the location data
#Reference grid : input parameters
RESO <- 100 # grid resolution (m)
BUFF <- 5000 # grid extent (m) (buffer around location extremes)
XMIN <- RESO*(round(((min(coords$X)-BUFF)/RESO),0))
YMIN <- RESO*(round(((min(coords$Y)-BUFF)/RESO),0))
XMAX <- XMIN+RESO*(round(((max(coords$X)+BUFF-XMIN)/RESO),0))
YMAX <- YMIN+RESO*(round(((max(coords$Y)+BUFF-YMIN)/RESO),0))
NRW <- ((YMAX-YMIN)/RESO)
NCL <- ((XMAX-XMIN)/RESO)
#Generation of refgrid
refgrid<-raster(nrows=NRW, ncols=NCL, xmn=XMIN, xmx=XMAX, ymn=YMIN, ymx=YMAX)
refgrid<-as(refgrid,"SpatialPixels")
#LKDE computation
ud <- kernelUD(loc, grid=refgrid, h="href")
# Volume contours computation
udvol1<-getvolumeUD(ud, standardize = FALSE)
#writeAsciiGrid(udvol, paste(substr(List[i,],1,7),"LKDE","asc", sep="."))
if (require(rgl)) {
#data(loc)
r1 <- raster(udvol1)
extent(r1) <- c(0, 610, 0, 870)
drape <- cut(r1, 5)
plot3D(r1, drape=drape, zfac=2)
}}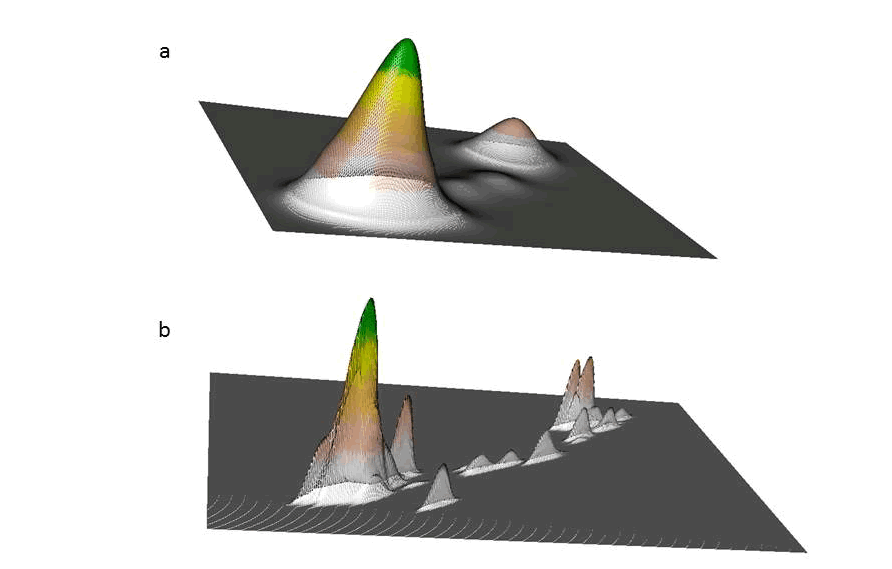
Figure 6.1: Example home range of a mule deer in 3D using KDE with a) href and b) hplug-in bandwidth selection. -
We can also run code for creating BBMM computation for further comparison
for(i in 1:nrow(List)) {coords<-read.table(as.character(List[i,]),sep="\t",header=T)
head(coords)loc<-coords[,c("X", "Y")]
coordinates(loc) = c("X", "Y")
#Coordinate system info may not be needed
proj4string(loc) = CRS("+proj=utm +zone=12 +datum=WGS84")
#Generation of a reference grid around the location data
#Reference grid : input parameters
RESO <- 100 # grid resolution (m)
BUFF <- 5000 # grid extent (m) (buffer around location extremes)
XMIN <- RESO*(round(((min(coords$X)-BUFF)/RESO),0))
YMIN <- RESO*(round(((min(coords$Y)-BUFF)/RESO),0))
XMAX <- XMIN+RESO*(round(((max(coords$X)+BUFF-XMIN)/RESO),0))
YMAX <- YMIN+RESO*(round(((max(coords$Y)+BUFF-YMIN)/RESO),0))
NRW <- ((YMAX-YMIN)/RESO)
NCL <- ((XMAX-XMIN)/RESO)
#Generation of refgrid
refgrid<-raster(nrows=NRW, ncols=NCL, xmn=XMIN, xmx=XMAX, ymn=YMIN, ymx=YMAX)
##Get the center points of the mask raster with values set to 1
refgrid <- xyFromCell(refgrid, 1:ncell(refgrid))
#BBMM computation
BBMM <- brownian.bridge(x=coords$X, y=coords$Y, time.lag=coords$timelag[-1], area.grid=refgrid, location.error=22, max.lag=18000)#Volume contours computation
#Create a data frame from x,y,z values
BBMM.df <- data.frame("x"=BBMM$x,"y"=BBMM$y,"z"=BBMM$probability)
##Make a raster from the x, y, z values, assign projection from above,
##match the resolution to that of the raster mask, note 100 is the cell
##resolution defined in evalPoints above
bbmm.raster <- rasterFromXYZ(BBMM.df, res=c(100,100), crs=proj4string(loc))
##Cast the data over to an adehabitatHR estUD
bbmm.px <- as(bbmm.raster, "SpatialPixelsDataFrame")
bbmm.ud <- new("estUD",bbmm.px)
bbmm.ud@vol = FALSE
bbmm.ud@h$meth = "BBMM"
##Convert the raw UD values to volume
udvol2 <- getvolumeUD(bbmm.ud, standardize=TRUE)
proj4string(udvol2) = CRS("+proj=utm +zone=12 +datum=WGS84")
if (require(rgl)) {
r2 <- raster(udvol2)
plot3D(r2,zfac=-2, xlim = XMIN, ylim = YMIN,xlab = "x", ylab = "y", zlab = "z",
rev=TRUE)
title3d('UD with Brownian Bridge Movement Model ')
decorate3d()
}}

