First we will begin with determining the distance between several features. In our first example, we want to measure distance from each mule deer location to the nearest stream if it is determined a priori that water or riparian habitats influence mule deer distribution in our study area. While this may not seem like a very complicated process, there are numerous steps needed to achieve this feat. We will need to use the package spatstat that will help us in creating individual segments with nodes for linear features such as roads and streams/rivers.
- Exercise 8.2 - Download and extract zip folder into your preferred location
- Set working directory to the extracted folder in R under File - Change dir...
- First we need to load the packages needed for the exercise
library(raster)
library(sp)
library(rgdal)
library(lattice)
library(rgeos)
library(spatstat) - Now open the script "LinearDistScript.R" and run code directly from the script
muleys <-read.csv("muleysexample.csv", header=T)
summary(muleys$id)
#Remove outlier locations
newmuleys <-subset(muleys, muleys$Long > -110.90 & muleys$Lat > 37.80)
muleys <- newmuleys
newmuleys <-subset(muleys, muleys$Long < -107)
muleys <- newmuleys
#Make a spatial data frame of locations after removing outliers
coords<-data.frame(x = muleys$X, y = muleys$Y)
crs<-"+proj=utm +zone=12 +datum=NAD83 +units=m +no_defs +ellps=GRS80 +towgs84=0,0,0"
#Convert to a SpatialPointsDataFrame
deer.spdf <- SpatialPointsDataFrame(coords= coords, data = muleys,
proj4string = CRS(crs))
deer.spdf[1:5,]
class(deer.spdf)
proj4string(deer.spdf) - Load the necessary road and rivers shapefiles already in Albers projection to match previous vegetation raster.
roads<-readOGR(dsn=".",layer="AlbersRoads")
rivers<-readOGR(dsn=".",layer="AlbersRivers")
plot(roads,pch=16)
points(deer.spdf, col="red")
plot(rivers,add=T, col="blue",pch=16) - We need to project the deer.spdf to Albers to match other layers
Albers.crs <-CRS("+proj=aea +lat_1=29.5 +lat_2=45.5 +lat_0=23 +lon_0=-96 +x_0=0 +y_0=0 +datum=NAD83 +units=m +no_defs +ellps=GRS80 +towgs84=0,0,0")
deer.albers <-spTransform(deer.spdf, CRS=Albers.crs)
points(deer.albers, col="red")
class(deer.albers)
proj4string(deer.albers)
deer.spdf[1:5,]
deer.albers[1:5,] - Determine boundary box around mule deer locations to create a layer to clip and zoom in.
bbox(deer.albers)
bb1 <- cbind(x=c(-1106865,-1106865,-1145027,-1145027, -1106865), y=c(1695607, 1729463,1729463,1695607,1695607))
AlbersSP <- SpatialPolygons(list(Polygons(list(Polygon(bb1)),"1")), proj4string=CRS(proj4string(deer.albers)))
plot(AlbersSP) - Load vegetation raster layer tif that came in the Albers projection from the online source.
crops <-raster("crop2012utm12.tif")
#Check to see all our layers are now in Albers projection
proj4string(crops)
proj4string(deer.albers)
proj4string(AlbersSP)
plot(crops)
points(deer.albers, col="red") - Clip the raster using the bounding box (AlbersSP) created in step 5.
bbclip <- crop(crops, AlbersSP) #Start re-run of code here after new clip#########
cliproads <- gIntersection(roads, AlbersSP, byid=TRUE)
cliprivers <- gIntersection(rivers, AlbersSP, byid=TRUE)
#Plot all to see if it is working for us and zoomed in to mule deer locations.
plot(bbclip)
points(deer.albers, col="red")
plot(cliproads, add=T)
plot(cliprivers, col="blue",add=T)
- Let's start with the road layer by converting a single line to a set of segments packaged as a function.
foo <- function(cliproads){
x <- cliproads@Lines[[1]]@coords
cbind(
head(x,-1),
tail(x,-1))}
#The function can be applied successively to each line in the list we extracted from roads. Results are output as a list, then converted to a matrix.
segs.lst <- lapply(cliproads@lines,foo)
segs <- do.call(rbind,segs.lst)
segs.x <- c(segs[,c(1,3)])
segs.y <- c(segs[,c(2,4)])
segs.owin <- as.owin(c(range(segs.x),range(segs.y)))
#The segments as a planar segment pattern:
segs.psp <- as.psp(segs, window=segs.owin)
plot(segs.psp)
points(deer.albers)
segs.psp
lengths.psp(segs.psp)
#We can cut road segments into lengths we control as well
dist <- pointsOnLines(segs.psp, eps=1000) - We need to back up a moment to handle the mule deer locations. Need to convert deer.albers from SPDF back to a dataframe because we need xy coordinates to be in Albers. NOTE: If all data is in UTM 12N or a similar projection from the beginning then no need for the next step. Then we need to make mule deer xy coordinates a planar point patter (i.e., ppp) for use in package spatstat.
deer2 <-as.data.frame(deer.albers)
deer2[1:5,]
newdeer <-cbind(deer2$x,deer2$y)
newdeer[1:5,]
xy.ppp <- as.ppp(newdeer,W=bdy)
plot(xy.ppp) - Also we need to back up and make the bounding polygon (AlbersSP) a class owin in order to proceed with functions in package spatstat.
AlbersSPDF <- as(AlbersSP, "SpatialPolygonsDataFrame")
#poly <- AlbersSPDF@polygons[[1]]@Polygons[[1]]@coords#These 2 lines won't work anymore due to gpclib issues
#bdy.gpc <- as(poly, "gpc.poly")
bdy.gpc <- as(AlbersSPDF, "gpc.poly")
bdy.owin <- gpc2owin(bdy.gpc)#new code using polyCub package
bdy <- as.polygonal(bdy.owin)
xy.ppp <- as.ppp(newdeer,W=bdy)
plot(xy.ppp)
#Let's check to determine if mule deer locations, bounding box, and road layer are in the proper classes to proceed.
is.owin(bdy.owin)
#[1] TRUE
is.ppp(xy.ppp)
#[1] TRUE
is.psp(segs.psp)
#[1] TRUE
#All is TRUE so now we can move forward with the analysis - Now we can determine the distance from mule deer locations (xy.ppp) to the nearest road
roaddist <- nncross(xy.ppp, segs.psp)$dist
roaddist
#Or identify segment number closest to each point
v <- nearestsegment(xy.ppp,segs.psp)#Identifies segment number not a distance
plot(segs.psp)
plot(xy.ppp[7501], add=TRUE, col="red")
plot(segs.psp[v[7501]], add=TRUE, lwd=5, col="red") - Now we do the same to a river layer by converting a single line to a set of segments packaged as a function.
foo <- function(cliprivers){
x <- cliprivers@Lines[[1]]@coords
cbind(
head(x,-1),
tail(x,-1))}
#Again, the function is applied successively to each line in the list then results are output as a list, then converted to a matrix.
rivs.lst <- lapply(cliprivers@lines,foo)
rivs <- do.call(rbind,rivs.lst)
rivs.x <- c(rivs[,c(1,3)])
rivs.y <- c(rivs[,c(2,4)])
rivs.owin <- as.owin(c(range(rivs.x),range(rivs.y)))
#The segments as a planar segment pattern:
rivs.psp <- as.psp(rivs, window=rivs.owin)
plot(rivs.psp)
points(deer.albers)
is.psp(rivs.psp)
#[1] TRUE
#All is TRUE so now we can move forward with the analysis - Now we can determine the distance from mule deer locations (xy.ppp) to the nearest river.
rivdist <- nncross(xy.ppp, rivs.psp)$dist
#rivdist #activate this code to see all distances
#Or identify segment number closest to each point
riv <- nearestsegment(xy.ppp,rivs.psp)
plot(rivs.psp, lwd=1)
plot(xy.ppp[1], add=TRUE, col="red")
plot(rivs.psp[riv[1]], add=TRUE, lwd=5, col="red")
#This code allows us to determine the nearest river to each deer location
plot(xy.ppp[290], add=TRUE, col="blue")
plot(rivs.psp[riv[290]], add=TRUE, lwd=5, col="blue")
- We can then summarize the distances in some meaningful way for analysis. Instead of representing distance to road as individual numerical values we can bin the distances in some categories we determine appropriate for our research objective.
br <- seq(0,4000,500)
lbl <- paste(head(br,-1),tail(br,-1),sep="-")
road.tbl <- table(cut(roaddist,breaks=br,labels=lbl))
Rdresults <- road.tbl/sum(road.tbl)
Rdresults
br1 <- seq(0,4000,500)
lbl1 <- paste(head(br1,-1),tail(br1,-1),sep="-")
river.tbl <- table(cut(rivdist,breaks=br1,labels=lbl1))
Rivresults <- river.tbl/sum(river.tbl)
Rivresults
#Or we can place each distance into a category or Bin for each deer.
library(Rcmdr)
BinRoad <- bin.var(roaddist, bins=5, method='intervals', labels=c('1','2','3','4','5'))
BinRoad
BinRivers <- bin.var(rivdist, bins=5, method='intervals', labels=c('1','2','3','4','5'))
BinRoad #end re-run here##########
#Let's try to add these distance covariates back to the original muley dataset.
Dist <- cbind(BinRoad,BinRivers)
muleys <- cbind(muleys, Dist)#Will not work because 2 locations are out of the box - We need to create a new boundary box that encompasses all location so as not to miss those 2. We can do this by zooming into the raster around all of the mule deer locations using a clever function in raster.
plot(veg)
points(deer.albers, col="red")
e <- drawExtent()
#click on top left of crop box and bottom right of crop box create zoom
bbox(deer.albers) #use these coordinates to create a boundary box
bb2 <- cbind(x=c(-1097405,-1097405,-1154795,-1154795, -1097405), y=c(1686777, 1739607,1739607,1686777,1686777))
AlbersSP <- SpatialPolygons(list(Polygons(list(Polygon(bb2)),"1")), proj4string=CRS(proj4string(deer.albers)))
plot(AlbersSP) - Now re-run code from clipping roads/streams to BinRoad and use cbind function to add binned distances to muleys dataset.
Dist <- cbind(BinRoad,BinRivers)
muleys <- cbind(muleys, Dist)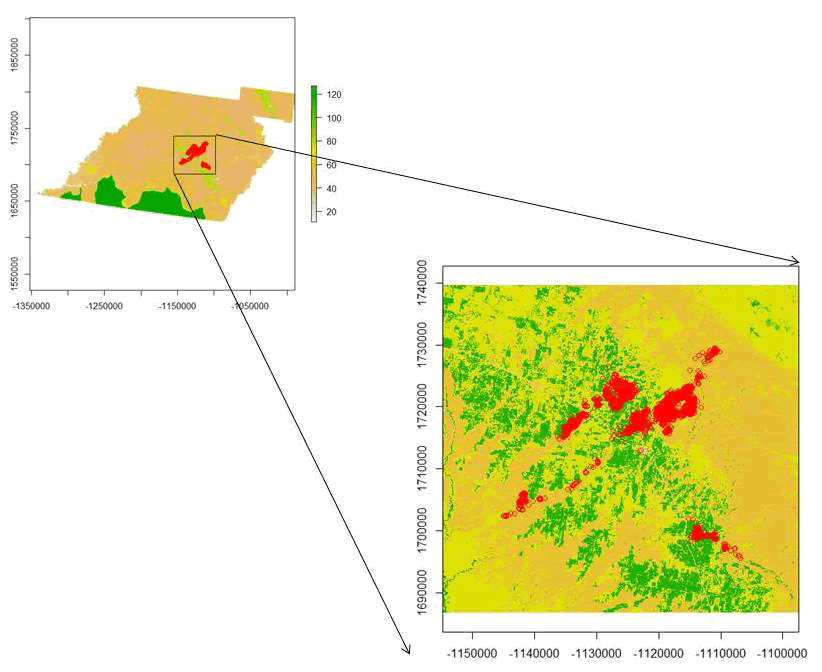
Figure 8.3: Zooming in around mule deer locations using drawExtent in the raster package in R.

