Using hplug-in in ks, we were able to calculate KDEs for the sample GPS datasets on 3 avian species and 2 mammalian species where first generation methods (hlscv) failed or generated a considerably over-smoothed KDE (href). While home range polygons generated with hplug-in appeared fragmented, they may be appropriate when studying a species in highly fragmented landscapes such as urban areas. Based on our results and previous research, conclusions presented in Loader 1999 should be re-evaluated for analyses of large GPS dataset because sample size and clumping of locations has consistently failed using hlscv while estimates using hplug-in converged for large multimodal datasets and resulted in reasonable estimates (Girard et al. 2002, Amstrup et al. 2004, Gitzen et al. 2006).
- Exercise 4.3 - Download and extract zip folder into your preferred location
- Set working directory to the extracted folder in R under File - Change dir...
- First we need to load packages needed for this exercise
library(ks)
library(rgdal)
library(maptools)
library(gpclib)
library(PBSmapping) - Now open the script "PelicanPlug.R" and run code directly from the script
- We will use an abbreviated dataset to save processing time and the code will also output shapefiles of home ranges
#Get input file
indata <- read.csv("White10pelicans.csv")
innames <- unique(indata$ID)
innames <- innames[1:5]
outnames <- innames - We then want to set up a table to output estimates of size of home ranges
#Set up output table
output <- as.data.frame(matrix(0,nrow=length(innames),ncol=9))
colnames(output) <- c("ID","noFixes","h11","h12","h21","h22", "iso50areaKm","iso95areaKm","iso99areaKm")
NOTE: the h followed by a number are outputs from the hplug-in for the bandwidth matrix estimated for each animal. They are in there for reference but don't really need them. - We also need to tell R which contours to output
#Set up levels for home range
levels <- c(50,95,99) - Set up a directory to place the resulting shapefiles
#Set up output directory for shp files
dir <- "output"
#Begin loop to calculate home ranges
for (i in 1:length(innames)){
data <- indata[which(indata$ID==innames[i]),]
if(dim(data)[1] != 0){
# export the point data into a shp file
data.xy = data[c("Longitude", "Latitude")]
coordinates (data.xy) <- ~Longitude+Latitude
sppt <- SpatialPointsDataFrame(coordinates(data.xy),data)
proj4string(sppt) <- CRS("+proj=longlat +ellps=WGS84")
writePointsShape(sppt,fn=paste(dir,outnames[i],sep="/"), factor2char=TRUE)
# start populating output table by column heading "pelicanID"
output$ID[i] <- data$ID[1]
output$noFixes[i] <- dim(data)[1]
locs <- cbind(data$Longitude,data$Latitude)
try(HpiOut <- Hpi(locs,pilot="samse",binned=TRUE))
if(is.null(HpiOut)=="FALSE"){
output$h11[i] <- HpiOut[1,1]
output$h12[i] <- HpiOut[1,2]
output$h21[i] <- HpiOut[2,1]
output$h22[i] <- HpiOut[2,2]
fhatOut <- kde(x=locs,H=HpiOut)
}
if(is.null(fhatOut)=="FALSE"){
for (j in 1:length(levels)){
fhat.contlev <- contourLevels(fhatOut, cont=c(levels[j]))
fhat.contlines <- contourLines(x=fhatOut$eval.points[[1]],
y=fhatOut$eval.points[[2]], z=fhatOut$estimate, level=fhat.contlev)
# convert contour lines into spatial objects to export as
polygon shp file
sldf <- ContourLines2SLDF(fhat.contlines)
proj4string(sldf) <- CRS("+proj=longlat +ellps=WGS84")
ps <- SpatialLines2PolySet(sldf)
attr(ps,"projection") <- "LL"
sp <- PolySet2SpatialPolygons(ps)
dataframe <- as.data.frame(matrix(as.character(1,nrow=1,ncol=1)))
spdf <- SpatialPolygonsDataFrame(sp,dataframe,match.ID=TRUE)
# get area and export shp files
if (j == 1){
pls <- slot(spdf, "polygons")[[1]]
gpclibPermit()
xx <- checkPolygonsHoles(pls)
a <- sapply(slot(xx, "Polygons"), slot, "area")
h <- sapply(slot(xx, "Polygons"), slot, "hole")
output$iso50areaKm[i] <- sum(ifelse(h, -a, a))/1000000
writeOGR(spdf,dirout,paste(outnames[i],"KUD50",sep=""), "ESRI Shapefile")}
if (j == 2){
pls <- slot(spdf, "polygons")[[1]]
gpclibPermit()
xx <- checkPolygonsHoles(pls)
a <- sapply(slot(xx, "Polygons"), slot, "area")
h <- sapply(slot(xx, "Polygons"), slot, "hole")
output$iso95areaKm[i] <- sum(ifelse(h, -a, a))/1000000
writeOGR(spdf,dirout,paste(outnames[i],"KUD95",sep=""),"ESRI Shapefile")}
if (j == 3){
pls <- slot(spdf, "polygons")[[1]]
gpclibPermit()
xx <- checkPolygonsHoles(pls)
a <- sapply(slot(xx, "Polygons"), slot, "area")
h <- sapply(slot(xx, "Polygons"), slot, "hole")
output$iso99areaKm[i] <- sum(ifelse(h, -a, a))/1000000
writeOGR(spdf,dirout,paste(outnames[i],"KUD99",sep=""), "ESRI Shapefile")}
}}}
rm(data,data.xy,sppt,locs,HpiOut,fhatOut,fhat.contlev, fhat.contlines,
sldf,ps,sp,dataframe,spdf)
}
#write output
write.csv(output,paste(dir,"\\output.csv",sep="")) - There is also an alternative way to create KDE hplug-in without using the looping environment in the code above. This also uses the ks package in a more straight forward manner (Duong and Hazelton 2003, Duong 2007).
- Exercise 4.3a - Download and extract zip folder into your preferred location
- Set working directory to the extracted folder in R under File - Change dir...
- First we need to load the packages needed for the exercise
library(ks)
library(rgdal)
library(maptools)
library(gpclib)
library(PBSmapping)
library(adehabitat)
library(adehabitatHR)
library(raster) -
Now open the script "PantherKSplug.R" and run code directly from the script
#Load dataset
panther<-read.csv("pantherjitter.csv",header=T)
str(panther)
panther$CatID <- as.factor(panther$CatID)
cat143 <- subset(panther, panther$CatID == "143")
##Get only the coordinates
loc <- data.frame("x"=cat143$X, "y"=cat143$Y)
##Define the projection of the coordinates
proj4string <- CRS("+proj=utm +zone=17N +ellps=WGS84")
##Make SpatialPointsDataFrame using the XY, attributes, and projection
spdf <- SpatialPointsDataFrame(loc, cat143, proj4string = proj4string)
81
#Calculate the bandwidth matrix to use later in creating the KDE
Hpi1 <- Hpi(x = loc)
Hpi1
##write out the bandwidth matrix to a file as you might want to refer to it later
#write.table(Hpi1, paste("hpivalue_", "143", ".txt", sep=""), row.names=FALSE,
#sep="\t")##Create spatial points from just the xyâAZs
loc.pts <- SpatialPoints(loc, proj4string=proj4string)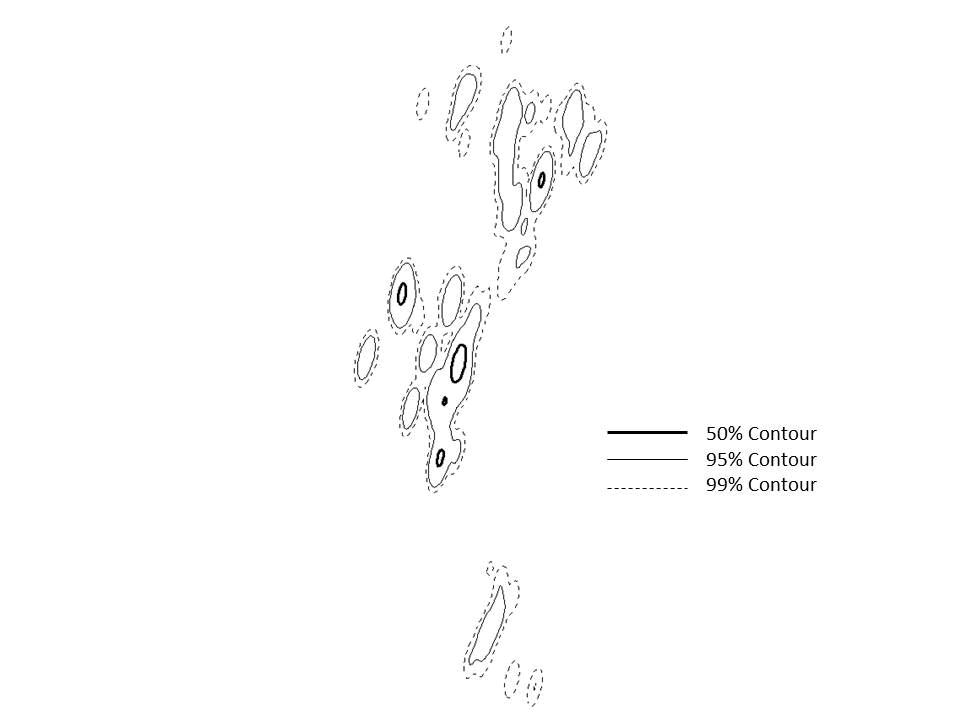
Figure 4.1: Example of KDE with hplug-in smoothing parameter to estimate size of home range for an American White Pelican. -
For home range calculations, some packages require evaluation points (ks) while others
require grid as spatial pixels (adehabitatHR). Understanding these simple concepts of
what formats of data are required by each package will save a tremendous amount of
time as we move forward!
##Set the expansion value for the grid and get the bbox from the
##SpatialPointsDataFrame
expandValue <- 2500 #This value is the amount to add on each side of the bbox
#Change to 5000 if error occurs at 99% ud
boundingVals <- spdf@bbox
##Get the change in x and y and adjust using expansion value
deltaLong <- as.integer(((boundingVals[1,2]) - (boundingVals[1,1]))
+ (2*expandValue))
deltaLat <- as.integer(((boundingVals[2,2]) - (boundingVals[2,1]))
+ (2*expandValue))
##100 meter grid for data in this exercise
gridRes <- 100
gridSizeX <- deltaLong / gridRes
gridSizeY <- deltaLat / gridRes
##Offset the bounding coordinates to account for the additional area
boundingVals[2,1] <- boundingVals[2,1] - expandValue
boundingVals[2,2] <- boundingVals[2,2] + expandValue
boundingVals[1,1] <- boundingVals[1,1] - expandValue
boundingVals[1,2] <- boundingVals[1,2] + expandValue
##Grid Topology object is basis for sampling grid (offset, cellsize, dim)
gridTopo <- GridTopology((boundingVals[,1]),
c(gridRes,gridRes),c(gridSizeX,gridSizeY))
##Using the Grid Topology and projection create a SpatialGrid class
sampGrid <- SpatialGrid(gridTopo, proj4string = proj4string)
##Cast over to Spatial Pixels
sampSP <- as(sampGrid, "SpatialPixels")
##convert the SpatialGrid class to a raster
sampRaster <- raster(sampGrid)
##set all the raster values to 1 such as to make a data masksampRaster[] <- 1
##Get the center points of the mask raster with values set to 1
evalPoints <- xyFromCell(sampRaster, 1:ncell(sampRaster))
##Here we can see how grid has a buffer around the locations and trajectory.
##This will ensure that we project our home range estimates into a slightly
##larger extent than the original points extent (bbox) alone.
plot(sampRaster)
#lines(tele.range.ltraj.lines, cex=0.5, lwd=0.1, col="grey")
points(loc, pch=1, cex=0.5)
##Create the KDE using the evaluation points
hpikde <- kde(x=loc, H=Hpi1, eval.points=evalPoints)
##Create a template raster based upon the mask and then assign the values from
##the kde to the template
hpikde.raster <- raster(sampRaster)
hpikde.raster <- setValues(hpikde.raster,hpikde$estimate)
##Lets take this raster and put it back into an adehabitat object
##This is convenient to use other adehabitat capabilities such as overlap
## indices or percent volume contours
##Cast over to SPxDF
hpikde.px <- as(hpikde.raster,"SpatialPixelsDataFrame")
##create new estUD using the SPxDF
hpikde.ud <- new("estUD", hpikde.px)
##Assign values to a couple slots of the estUD
hpikde.ud@vol = FALSE
hpikde.ud@h$meth = "Plug-in Bandwidth"
##Convert the UD values to volume using getvolumeUD from adehabitatHR
##and cast over to a raster
hpikde.ud.vol <- getvolumeUD(hpikde.ud, standardize=TRUE)
hpikde.ud.vol.raster <- raster(hpikde.ud.vol)
##Here we generate volume contours using the UD
hpikde.50vol <- getverticeshr(hpikde.ud, percent = 50,ida = NULL, unin = "m", unout = "ha", standardize=TRUE)
hpikde.80vol <- getverticeshr(hpikde.ud, percent = 80,ida = NULL, unin = "m", unout = "ha", standardize=TRUE)
hpikde.90vol <- getverticeshr(hpikde.ud, percent = 90,ida = NULL, unin = "m",
unout = "ha", standardize=TRUE)
hpikde.95vol <- getverticeshr(hpikde.ud, percent = 95,ida = NULL, unin = "m", unout = "ha", standardize=TRUE)
hpikde.99vol <- getverticeshr(hpikde.ud, percent = 99,ida = NULL, unin = "m", unout = "ha", standardize=TRUE)##Let's put the HR, volume, volume contours, and points on a plot
##and write out the shapefiles for contours for use in ArcMap
plot.new()
breaks <- c(0, 50, 80, 90, 95, 99)
plot(hpikde.ud.vol.raster, col=heat.colors(3), breaks=breaks,interpolate=TRUE,
main="Kernel Density Estimation, Plug-in Bandwidth",xlab="Coords X",
ylab="Coords Y", legend.shrink=0.80,legend.args=list(text="UD by
Volume (%)",side=4, font=2, line=2.5, cex=0.8))
plot(hpikde.99vol, add=T)#col="grey",axes=T)
plot(hpikde.95vol, add=TRUE)
plot(hpikde.90vol, add=TRUE)
plot(hpikde.80vol, add=TRUE)
plot(hpikde.50vol, add=TRUE)
points(loc, pch=1, cex=0.5)
writeOGR(hpikde.50vol,dsn="output", layer="cat143plug50",driver="ESRI Shapefile", overwrite=TRUE)
writeOGR(hpikde.80vol,dsn="output", layer="cat143plug80",driver="ESRI Shapefile", overwrite=TRUE)
writeOGR(hpikde.90vol,dsn="output", layer="cat143plug90",driver="ESRI Shapefile", overwrite=TRUE)
writeOGR(hpikde.95vol,dsn="output", layer="cat143plug95",driver="ESRI Shapefile", overwrite=TRUE)
writeOGR(hpikde.99vol,dsn="output", layer="cat143plug99",driver="ESRI Shapefile", overwrite=TRUE)

