- Exercise 1.6 - Download and extract zip folder into your preferred location
- Set working directory to the extracted folder in R under File - Change dir...
- First we need to load the packages needed for the exercise
library(rgdal)
library(maptools)
library(foreign) - Now open the script "SoilScript.R" and run code directly from the script
soils<-readOGR(dsn=".",layer="Soil_Properties")
soils@proj4string ###get projection
plot(soils)
names(soils) ###get attribute data
#Rename ArcMap category headings to something more familiar
soils$Clay <- soils$SdvOutpu_1
soils$pH <- soils$SdvOutpu_2
soils$CEC <- soils$SdvOutpu_3
#Shapefiles contain several slots which can be called with the "@" symbol
#or slot(object, "data")
soils@data #= a data frame with n observations associated with X covariates,
soils@polygons #=the number of polygons that the shapefile consists of
soils@plotOrder #= the order of the polygons
soils@bbox #= boundary box
soils@proj4string #= projection
#Within the slot
soils@polygons [[1]] ###will bring up the first polygon
soils@polygons [[1]]@area ###will bring up the area for the first polygon
soils@polygons[[1]]@ID ##will retrieve the ID of the first polygon
soils@polygons[[1]]@plotOrder ##will retrieve the order of the first polygon - Select portions of the data that fit some set criteria
##Highlights the areas that Percent Clay polygons are over 30%
plot(soils, col=grey(1-soils$Clay > 30))
plot(soils)
high.clay<- soils[soils$Clay>30,]
plot(high.clay, border="red", add=TRUE)
##Highlights the areas that Cation Exchange Capacity is greater than 14
high.CEC<- soils[soils$CEC>14,]
plot(high.CEC, border="green", add=TRUE)
##Highlights the areas that soil pH is greater than 8
high.pH <- soils[soils$pH>8,]
plot(high.pH, border="yellow", add=TRUE) - Bring in locations of harvested mule deer
#Import mule deer locations from harvested animals tested for CWD
mule <-read.csv("MDclip.csv", header=T)
str(mule)
coords<-data.frame(x = mule$x, y = mule$y)
crs<-"+proj=utm +zone=13 +datum=WGS84 +no_defs +towgs84=0,0,0"
coords
plot(coords, col="blue")
par(new=TRUE) - Let's generate random points with the extent of the soil layer
#Sampling points in a Spatial Object### will give a regular grid
samples<-spsample(soils, n=1000, )
samples@proj4string
#Plot them to see if it worked or to create output figures
plot(soils, col="wheat")
points(coords, col="blue")
points(samples, col="red") - Creates a SpatialPoints object from the location coordinates
samples@bbox <- soils@bbox
samples@proj4string <- soils@proj4string - Extract and tally Clay soil types for random samples and mule deer locations:
#Match points with soil polygons they occur in
soils.idx<- over(samples,soils)
locs <- SpatialPoints(coords)
locs@proj4string <- soils@proj4string
soils.locs<- over(locs, soils)
#Tally clay soil types for random samples
obs.tbl <- table(soils.idx$Clay[soils.idx$Clay])
obs.tbl
#Also tally soil types for each mule deer sampled
obs.tbl2 <- table(soils.locs$Clay[soils.locs$Clay])
obs.tbl2 - Convert the counts to proportions:
obs <- obs.tbl/sum(obs.tbl)
obs
obs2 <- obs.tbl2/sum(obs.tbl2)
bs2
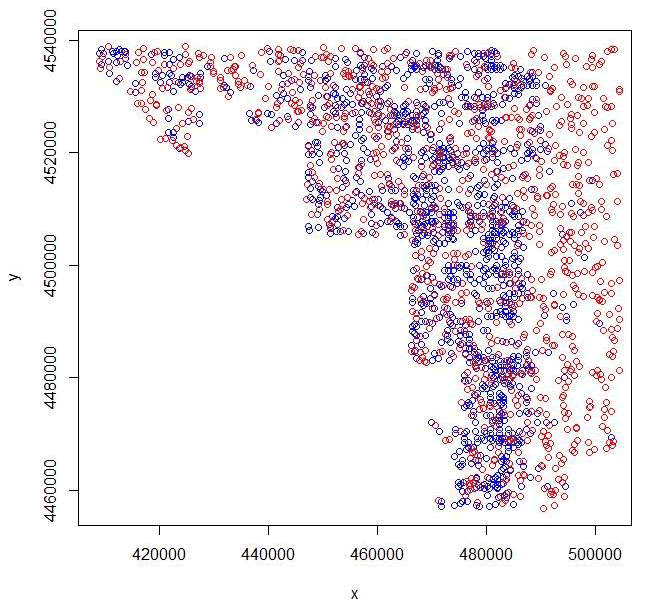
Figure 1.6 Overlay of mule deer locations with random locations generated in R

